Processing Geometric Imperfection Data¶
This part of the turorial will explain in a step-by-step example how one can use the developed modules to post-process measured imperfection data.
The following steps will be discussed:
raw data from measurement system
finding
Raw Data from Mesurement Systems¶
The raw files obtained from measured geometric imperfections usually come with three columns:
x1 y1 z1
x2 y2 z2
...
xn yn zn
The following sample has been used to illustrate this tutorial (first 10 lines shown below):
310.909 1174.136 261.8
146.126 754.321 378.119
216.694 1111.78 342.928
-261.395 1031.417 -314.052
-5.499 99.519 -407.764
-70.556 95.452 -401.672
-336.88 1148.365 -229.665
-351.651 705.414 204.226
403.703 454.238 -37.619
122.444 55.322 386.451
The following script is used to plot the sample with the imperfection exagerated. Note that best_fit_raw_data.py has to be executed before.
plot_sample_3d.py
import numpy as np
import mayavi.mlab as mlab
from desicos.conecylDB.read_write import xyz2thetazimp
R = 406.4 # nominal radius
H = 1219.2 # nominal height
R_best_fit = 407.193 # radius obtained with the best-fit routine
thetas, zfit, dR = np.genfromtxt('sample_theta_z_imp.txt', unpack=True)
xpos = R_best_fit*np.cos(thetas)
ypos = R_best_fit*np.sin(thetas)
sf = 20
x2 = xpos + sf*dR*np.cos(thetas)
y2 = ypos + sf*dR*np.sin(thetas)
z2 = zfit
Tinv = np.loadtxt('tmp_Tinv.txt')
x, y, z = Tinv.dot(np.vstack((x2, y2, z2, np.ones_like(x2))))
black = (0,0,0)
white = (1,1,1)
mlab.figure(bgcolor=white)
mlab.points3d(x, y, z, color=(0,1,0))
mlab.plot3d([0, 600], [0, 0], [0, 0], color=black, tube_radius=10.)
mlab.plot3d([0, 0], [0, 1600], [0, 0], color=black, tube_radius=10.)
mlab.plot3d([0, 0], [0, 0], [0, 600], color=black, tube_radius=10.)
mlab.text3d(650, -50, +50, 'x', color=black, scale=100.)
mlab.text3d(0, 1650, +50, 'y', color=black, scale=100.)
mlab.text3d(0, -50, 650, 'z', color=black, scale=100.)
mlab.savefig('plot_sample_3d.png', size=(500, 500))
giving:
In order to transform the raw data to the finite element coordinate
system, the recommended procedure is the function
xyz2thetazimp(), exemplified below. This function converts the “
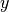
 ” data to a to convert them to a “
” data to a to convert them to a “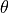

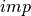 ” format. This function
is a wrapper that calls funcion
” format. This function
is a wrapper that calls funcion best_fit_cylinder() or
best_fit_cone(), where the right coordinate transformation is
calculated.
best_fit_raw_data.py
import numpy as np
from desicos.conecylDB.read_write import xyz2thetazimp
mps, out = xyz2thetazimp('sample.txt', alphadeg_measured=0, R_expected=406.4,
H_measured=1219.2, clip_bottom=10, clip_top=10, use_best_fit=True,
best_fit_output=True, rotatedeg=0.)
np.savetxt('tmp_Tinv.txt', out['Tinv'])
It will create an output file with a prefix "_theta_z_imp.txt" where the
imperfection data is stored in the format:
theta1 z1 imp1
theta2 z2 imp2
...
thetan zn impn
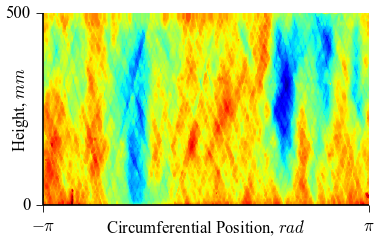
where thetai and zi are the coordinates 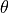 and
and  , while
, while
impi represents the imperfection amplitude at this coordinate. An example
of the transformed imperfection data is given below (first 10 lines), and
the full output is given here:
0.701704 44.752223 -0.299107
1.202029 464.737892 -0.772438
1.008550 107.201740 -0.834017
-2.266294 188.347008 0.697946
-1.581399 1119.953672 -0.574269
-1.742285 1124.099646 -0.728166
-2.545140 71.468489 0.131696
2.612788 514.312597 -0.417002
-0.090133 764.620654 -1.094824
1.261926 1163.763315 -0.307154
Note
It is also possible to use xyz2thetazimp() in other
ways, as detailed in the function’s documentation
The transformed data can also be visualized in the 3-D domain, which was accomplished using the following script:
plot_sample_3d_tranformed.py
import numpy as np
import mayavi.mlab as mlab
from desicos.conecylDB.read_write import xyz2thetazimp
R = 406.4
R_best_fit = 407.168
data = np.genfromtxt('sample_theta_z_imp.txt')
thetas, zfit, dR = data.T
xpos = R_best_fit*np.cos(thetas)
ypos = R_best_fit*np.sin(thetas)
sf = 20
x2 = xpos + sf*dR*np.cos(thetas)
y2 = ypos + sf*dR*np.sin(thetas)
z2 = zfit
black = (0,0,0)
white = (1,1,1)
mlab.figure(bgcolor=white)
mlab.points3d(x2, y2, z2, color=(0.5,0.5,0.5))
mlab.plot3d([0, 600], [0, 0], [0, 0], color=black, tube_radius=10.)
mlab.plot3d([0, 0], [0, 600], [0, 0], color=black, tube_radius=10.)
mlab.plot3d([0, 0], [0, 0], [0, 1600], color=black, tube_radius=10.)
mlab.text3d(650, -50, +50, 'x', color=black, scale=100.)
mlab.text3d(0, 650, +50, 'y', color=black, scale=100.)
mlab.text3d(0, -50, 1650, 'z', color=black, scale=100.)
mlab.savefig('plot_sample_3d_transformed.png')
giving:
Note that due to the axisymmetry it may happen that the final result has some
offset rotation. For this reason the interpolation routines that will be
discussed in the next sessions of this tutorial include a rotatedeg
parameter that the analyst should use in order to correct this angular offset.
Inverse-Weighted Interpolation¶
Function iterp_theta_z_imp() is the one used to interpolate the
measured data into a given mesh. In the example below the mesh creates using
np.meshgrid() is used for plotting purposes. Note that the an angular
offset of -88.5 degrees was used in order to adjust the imperfection.
interpolate_inv_weighted.py
import numpy as np
from desicos.conecylDB.interpolate import interp_theta_z_imp
R_model = 406.4 + 0.8128/2.
H_measured = 1219.2
ignore_bot_h = 25.4
ignore_top_h = 25.4
nz = 180
nt = 560
theta = np.linspace(-np.pi, np.pi, nt)
z = np.linspace(0, H_measured, nz)
T, Z = np.meshgrid(theta, z, copy=False)
data = np.loadtxt('sample_theta_z_imp.txt')
ts, zs, imps = data.T
a = np.vstack((ts, zs, imps)).T
mesh = np.vstack((T.ravel(), Z.ravel())).T
w0 = interp_theta_z_imp(data=a, mesh=mesh, semi_angle=0,
H_measured=H_measured, H_model=H_measured, R_model=R_model,
stretch_H=False, rotatedeg=-88.5, num_sub=100, ncp=5,
power_parameter=2, ignore_bot_h=ignore_bot_h,
ignore_top_h=ignore_top_h)
np.savetxt('tmp_inv_weighted.txt',
np.vstack((T.ravel(), Z.ravel(), w0)).T, fmt='%1.8f')
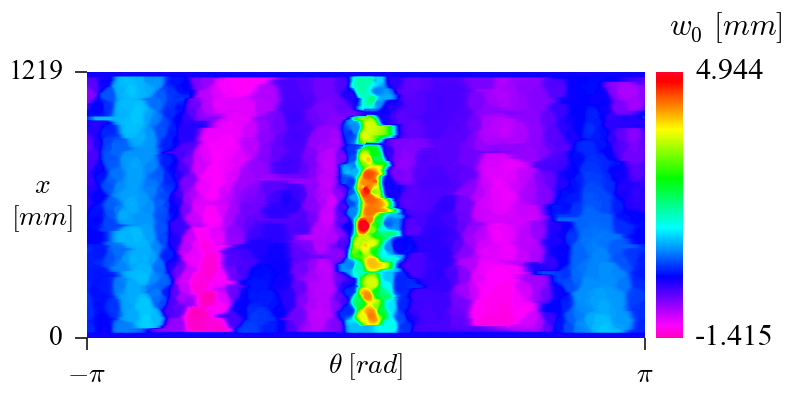
An the generated data can be plotted using the following code:
plot_inv_weighted.py
import numpy as np
from numpy import pi
import matplotlib.cm as cm
import matplotlib.pyplot as plt
plt.rcParams['axes.labelsize'] = 9.
plt.rcParams['font.size'] = 9.
path = 'tmp_inv_weighted.txt'
nz = 180
nt = 560
R = 407.168
#cmap = cm.jet
cmap = cm.gist_rainbow_r
theta, z, w0 = np.loadtxt(path, unpack=True)
fig = plt.figure(figsize=(3.5, 3.5), dpi=220)
levels = np.linspace(w0.min(), w0.max(), 400)
contour = plt.contourf(theta.reshape(nz, nt)*R, z.reshape(nz, nt),
w0.reshape(nz, nt), levels=levels, cmap=cmap)
ax = plt.gca()
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.set_aspect('equal')
ax.xaxis.set_ticks([-pi*R, pi*R])
ax.xaxis.set_ticklabels([r'$-\pi$', r'$\pi$'])
ax.set_xlabel(r'$\theta$ $[rad]$', ha='center', va='center')
ax.xaxis.labelpad=-7
ax.yaxis.set_ticks([0, 1219.2])
ax.set_ylabel('$x$\n$[mm]$', rotation='horizontal', ha='center',
va='center')
ax.yaxis.labelpad=-10
colorbar=True
if colorbar:
from mpl_toolkits.axes_grid1 import make_axes_locatable
fsize = 10
cbar_nticks = 2
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size='5%', pad=0.05)
cbarticks = np.linspace(w0.min(), w0.max(), cbar_nticks)
cbar = plt.colorbar(contour, ticks=cbarticks, format='%1.3f',
cax=cax, cmap=cmap)
cax.text(0.5, 1.1, '$w_0$ $[mm]$', ha='left', va='bottom',
fontsize=fsize)
cbar.outline.remove()
cbar.ax.tick_params(labelsize=fsize, pad=0., tick2On=False)
fig.savefig('plot_inv_weighted.png', transparent=True,
bbox_inches='tight', pad_inches=0.05, dpi=220)
giving:
Half-Cosine Function¶
Function calc_c0() is used to interpolate the measured data using
Fourier series. The half-cosine function corresponds to the parameter
funcnum = 2, as can be seen in the function’s documentation.
The following script was used to calculate the coefficients 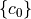 . Note
how the angular offset is applied using the
. Note
how the angular offset is applied using the rotatedeg parameter:
interpolate_half_cosine.py
import numpy as np
from desicos.conecylDB.fit_data import calc_c0
method = 'points'
funcnum= 2
path = 'sample_theta_z_imp.txt'
R = 406.4 + 0.8128/2.
H = 1219.2
m0 = 10
n0 = 10
c0, residues = calc_c0(path, m0=m0, n0=n0, funcnum=funcnum, rotatedeg=-88.5)
outname = 'tmp_c0_{0}_f{1:d}_m0_{2:03d}_n0_{3:03d}.txt'.format(method,
funcnum, m0, n0)
np.savetxt(outname, c0)
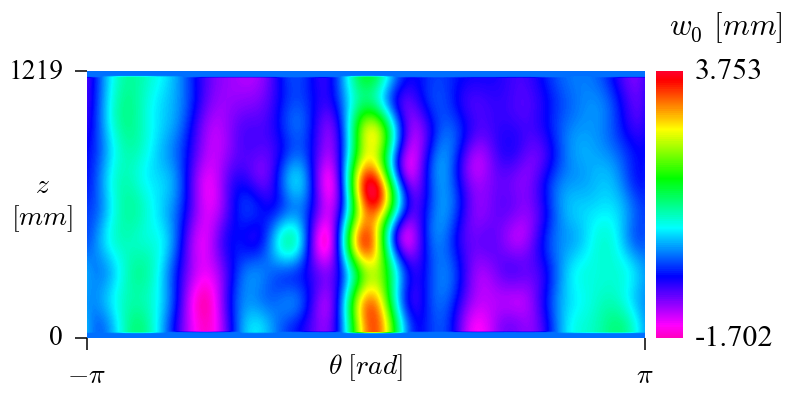
and the following routine was used to plot the corresponding imperfection field:
plot_half_cosine.py
import numpy as np
from numpy import pi
import matplotlib.cm as cm
import matplotlib.pyplot as plt
from compmech.conecyl.imperfections import fw0
plt.rcParams['axes.labelsize'] = 9.
plt.rcParams['font.size'] = 9.
method = 'points'
funcnum = 2
names = ['C01']
nz = 180
nt = 560
H = 1219.2
zpts_min = 25.4
zpts_max = H - 25.4
R = 406.4 + 0.8128/2.
#cmap = cm.jet
cmap = cm.gist_rainbow_r
m0 = 10
n0 = 10
filename = 'tmp_c0_{0}_f{1:d}_m0_{2:03d}_n0_{3:03d}.txt'.format(method,
funcnum, m0, n0)
c0 = np.loadtxt(filename)
ts = np.linspace(-pi, pi, nt)
xs = np.linspace(0, 1., nz)
zs = xs*H
check = (zs >= zpts_min) & (zs <= zpts_max)
test = xs[check]
xs -= test.min()
xs /= xs.max()
xs[xs<0] = 0
xs[xs>1] = 1
dummy, xs = np.meshgrid(ts, xs, copy=False)
ts, zs = np.meshgrid(ts, zs, copy=False)
w0 = fw0(m0, n0, c0, xs, ts, funcnum=funcnum)
w0.flat[(zs.flat < zpts_min) | (zs.flat > zpts_max)] = 0
fig = plt.figure(figsize=(3.5, 3.5), dpi=220)
levels = np.linspace(w0.min(), w0.max(), 400)
contour = plt.contourf(ts*R, zs, w0.reshape(ts.shape), levels=levels,
cmap=cmap)
ax = plt.gca()
ax.set_aspect('equal')
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['bottom'].set_visible(False)
ax.spines['top'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.xaxis.set_ticks([-pi*R, pi*R])
ax.xaxis.set_ticklabels([r'$-\pi$', r'$\pi$'])
ax.set_xlabel(r'$\theta$ $[rad]$', va='center', ha='center')
ax.xaxis.labelpad = -7
ax.yaxis.set_ticks([0, H])
ax.set_ylabel('$z$\n$[mm]$', rotation='horizontal', va='center',
ha='center')
ax.yaxis.labelpad = -10
colorbar = True
if colorbar:
from mpl_toolkits.axes_grid1 import make_axes_locatable
fsize = 10
cbar_nticks = 2
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size='5%', pad=0.05)
cbarticks = np.linspace(w0.min(), w0.max(), cbar_nticks)
cbar = plt.colorbar(contour, ticks=cbarticks, format='%1.3f',
cax=cax, cmap=cmap)
cax.text(0.5, 1.1, '$w_0$ $[mm]$', ha='left', va='bottom',
fontsize=fsize)
cbar.outline.remove()
cbar.ax.tick_params(labelsize=fsize, pad=0., tick2On=False)
fig.savefig('plot_half_cosine.png', transparent=True, bbox_inches='tight',
pad_inches=0.05, dpi=220)
giving:
The relatively poor correlation between the half-cosine and the inverse_weighted is primarily due to the reduced amount of measured data used for this tutorial. Using real imperfection data the correlation is much closer, as shown in the additional examples below.
Additional Examples using the Half-Cosine Function¶
The example below shows how to use this function and then plot the displacement patterns using different approximation levels:
half_cosine_additional_example.py
from numpy import *
from desicos.conecylDB.fit_data import fw0
ntheta = 420
nz = 180
funcnum = 2
path = 'degenhardt_2010_z25_msi_theta_z_imp.txt'
theta = linspace(-pi, pi, ntheta)
z = linspace(0., 1., nz)
theta, z = meshgrid(theta, z, copy=False)
for m0, n0 in [[20, 30], [30, 45], [40, 60], [50, 75]]:
c, residues = calc_c0(path, m0=m0, n0=n0, sample_size=(10*2*m*n),
funcnum=funcnum)
w0 = fw0(m0, n0, z.ravel(), theta.ravel(), funcnum=funcnum)
plt.figure(figsize=(3.5, 2))
levels = np.linspace(w0.min(), w0.max(), 400)
plt.contourf(theta, z*H_measured, w0.reshape(theta.shape),
levels=levels)
ax = plt.gca()
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.xaxis.set_ticks([-pi, pi])
ax.xaxis.set_ticklabels([r'$-\pi$', r'$\pi$'])
ax.set_xlabel('Circumferential Position, $rad$')
ax.xaxis.labelpad=-10
ax.yaxis.set_ticks([0, 500])
ax.set_ylabel('Height, $mm$')
ax.yaxis.labelpad=-15
filename = 'fw0_f{0}_z25_m_{1:03d}_n_{2:03d}.png'.format(
funcnum, m0, n0)
plt.gcf().savefig(filename, transparent=True, bbox_inches='tight',
pad_inches=0.05, dpi=90)
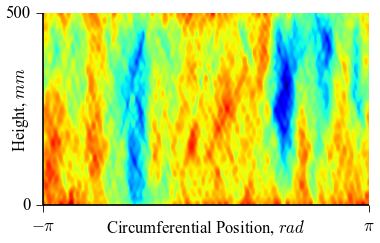
which will result in the following figures for funcnum=1:
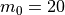 ,
, 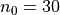 :
:
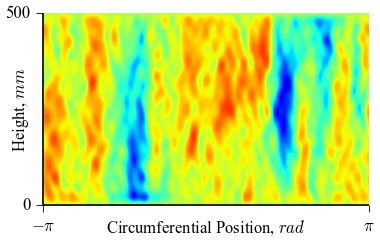
 ,
,  :
:
 ,
,  :
:
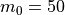 ,
, 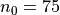 :
:
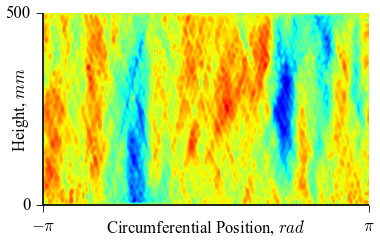
and for funcnum=2:
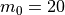 ,
, 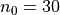 :
:
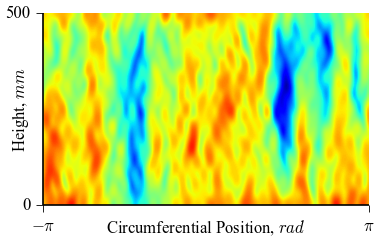
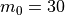 ,
, 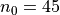 :
:
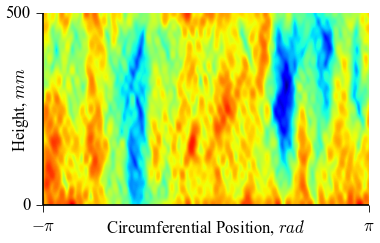
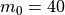 ,
, 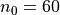 :
:
 ,
, 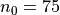 :
:
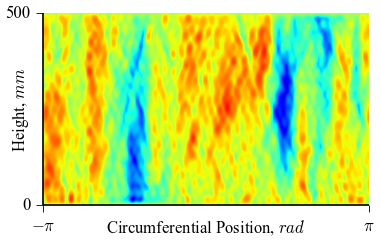
It can be seen how the  function approximates the measured imperfection
pattern with the increase of
function approximates the measured imperfection
pattern with the increase of  and
and 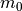 . The measured imperfection
pattern is shown below, and it was plotted after performing the
inverse-weighted interpolation already described.
. The measured imperfection
pattern is shown below, and it was plotted after performing the
inverse-weighted interpolation already described.
Comparing the results using funcnum=1 and funcnum=2, one see that
the latter approaches closer the real measurements, since it relaxes the
condition 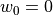 at the edges.
at the edges.
Processing Measured Thickness Imperfection Data¶
The procedure described for geometric imperfections is also applicable for
thickness imperfections. The measured imperfection data usually
comes in a 4 columns format where the thickness measurement is given for each
spatial position: 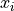 ,
, 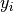 ,
, 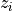 :
:
x1 y1 z1 thick1
x2 y2 z2 thick2
...
xn yn zn thickn
The recommended procedure to transform the raw data to the finite
element coordinate system is the function
xyzthick2thetazthick(), that also calls internally the
best_fit_cylinder() or the best_fit_cone() routines:
>>> from desicos.conecylDB.read_write import xyzthick2thetazthick
>>> xyzthick2thetazthick(path, alphadeg_measured, R_measured, H_measured,
fmt='%1.8f')
Note
It is possible to use function xyzthick2thetazthick`()
in other ways, as detailed in the corresponding documentation